Clement Chow

Assistant Professor of Human Genetics
Molecular Neuroscience
Neurobiology Disease
Developmental Neurobiology
Education
B.A. 2003, Cornell University; Ph.D. 2008; University of Michigan; Postdoctoral Fellow, 2008-2015, Cornell University
RESEARCH:
Genetic variation, Genomics, ER stress, Disease modifiers
Our lab is focused on understanding the role of genetic variation on disease outcomes. We employ quantitative and functional tools, in a variety of model organisms, to study how genetic variation impacts basic cellular traits important to human health. Our work in model organisms will help to model and inform studies of genetic variation in the human population. We hope to identify variation in the human population that can lead to more precise, personalized therapies.
The endoplasmic reticulum (ER) is a large organelle responsible for the synthesis, maturation, and delivery of proteins essential for cellular function. ER stress occurs when misfolded proteins accumulate in the ER lumen and if left unresolved, results in cell death. The cell responds to ER stress with the conserved "unfolded protein response" (UPR). The UPR returns the ER to homeostasis by attenuating protein synthesis, activating transcriptional signaling cascades, and refolding or degrading misfolded proteins in the ER.
ER stress can contribute to the pathogenesis of many different human diseases, like obesity, diabetes, and neurodegeneration. In many of these cases, genetic or pharmacological manipulation of ER stress levels influences disease outcome. This suggests that differing ER stress responses in patients may determine disease severity.
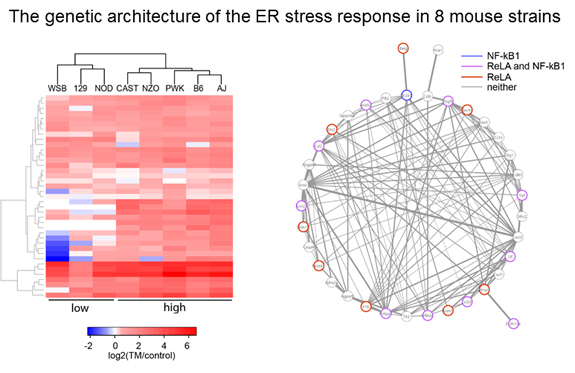
Genetic architecture of the ER stress response
Genetic variation is a powerful, unbiased tool to uncover novel aspects of gene networks.
We are using natural variation to understand how endoplasmic reticulum (ER) stress
varies among individuals in a population and what this means for disease variability.
To do this, we are using the mouse Collaborative Cross to uncover the genetic architecture
underlying this important cellular stress response. We are also utilizing Drosophila,
mouse, and cell culture studies to uncover the function of genes that contribute to
variability in the ER stress response.
Genomic adaptation to ER stress
In order to fully understand how genetic variation impacts the ER stress response,
we are also undertaking studies to deeply characterize the genomic response to ER
stress. How does the epigenome and the transcriptome adapt when a tissue or cell is
challenged with acute vs. prolonged ER stress? And more importantly, how do these
states differ in a tissue- or cell-specific manner? To do this we are using RNAseq
and various epigenetic tools to measure in vivo responses to ER stress in the mouse.
Mendelian diseases as quantitative traits
Natural genetic variation can also be used as a tool to identify novel modifiers of
Mendelian diseases. It is widely appreciated that the expressivity of many Mendelian
diseases can be quite variable. However, there is still a large gap in understanding
why this variability exists. We use natural genetic variation in model organisms to
study this variability in a controlled, tractable manner. We think that natural variation
(rather than mutagenesis) in model organisms better models genetic variation in the
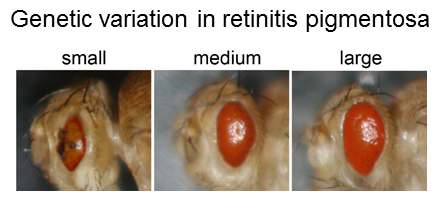
human population. As a first step, we tested a well-studied retinitis pigmentosa mutation
on ~200 strains of Drosophila and found extensive phenotypic variation in retinal
degeneration. Using association study techniques we identified and validated numerous
novel modifier genes. We are applying this paradigm to other rare Mendelian diseases,
like NGLY1 deficiency, to try to understand the nature of genetic modification.